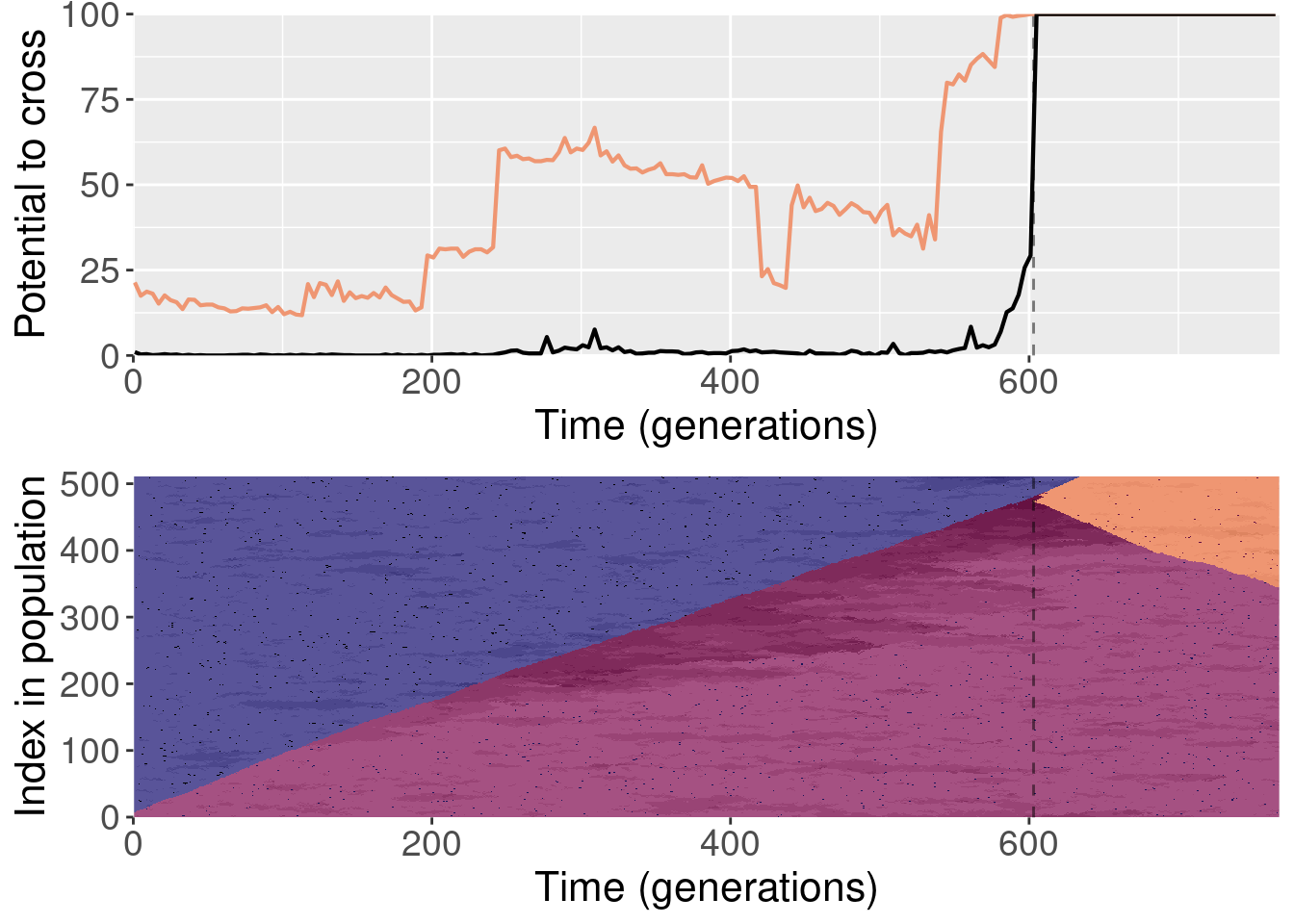
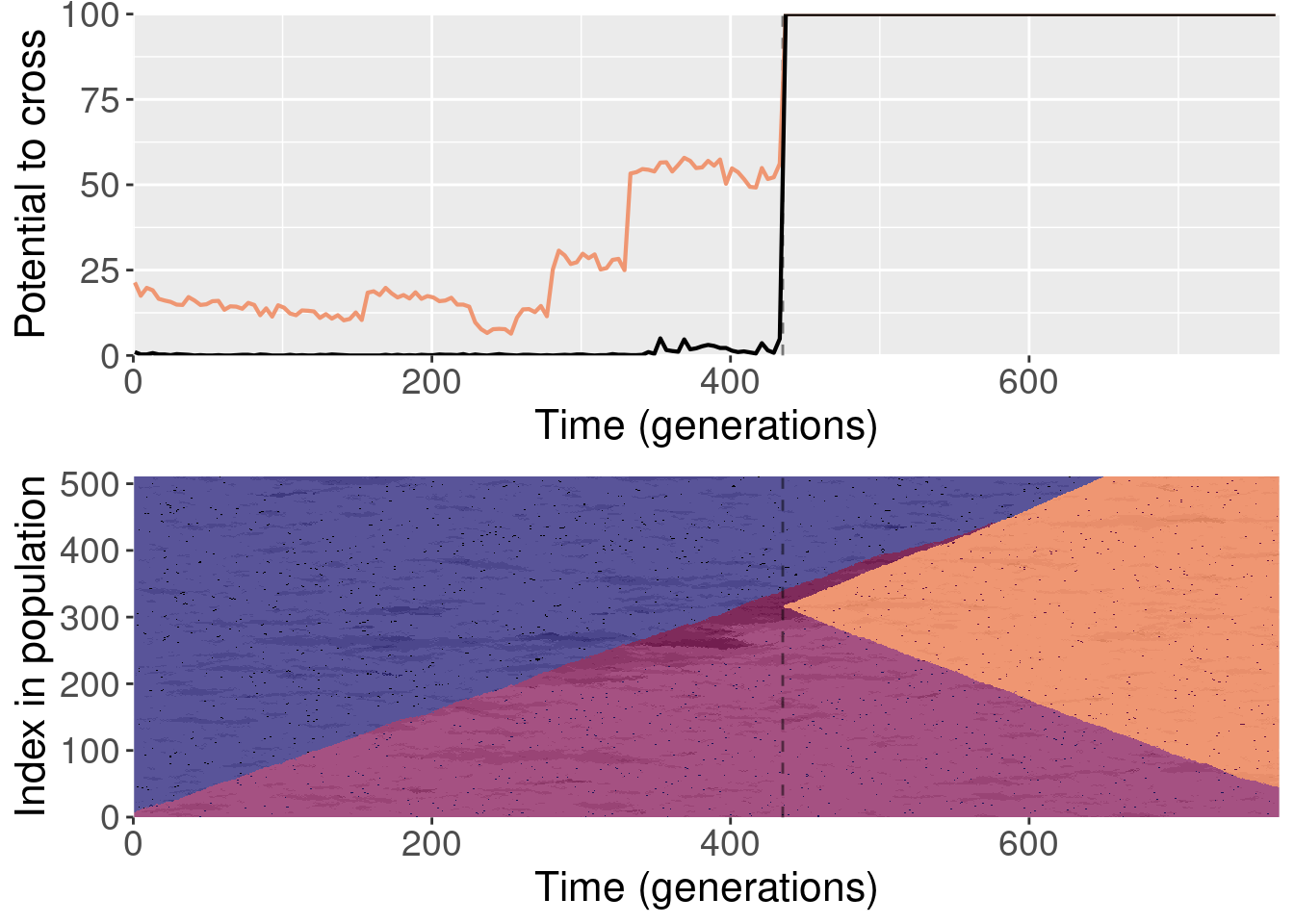
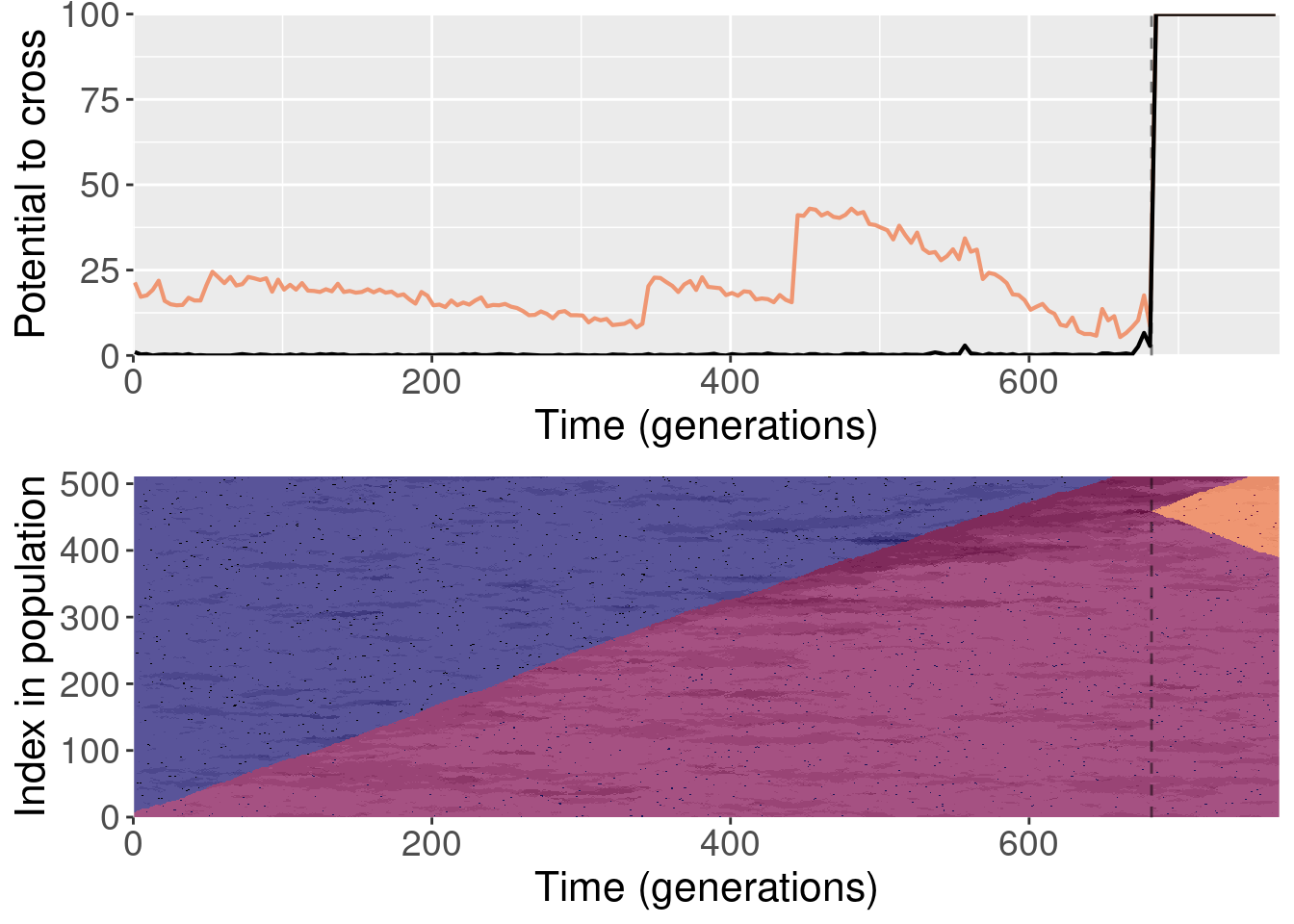
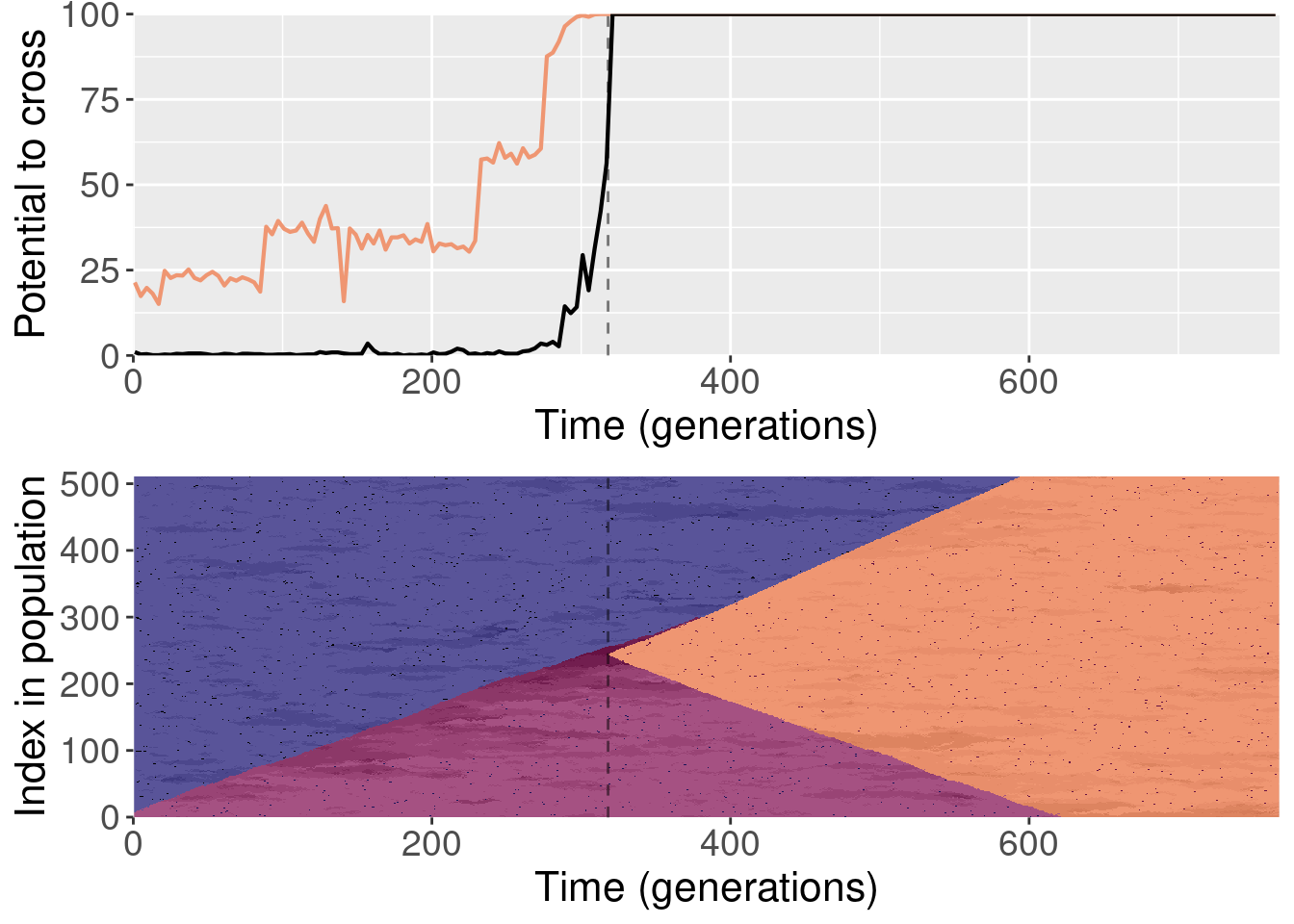
Section 4 Replaying shuffled populations
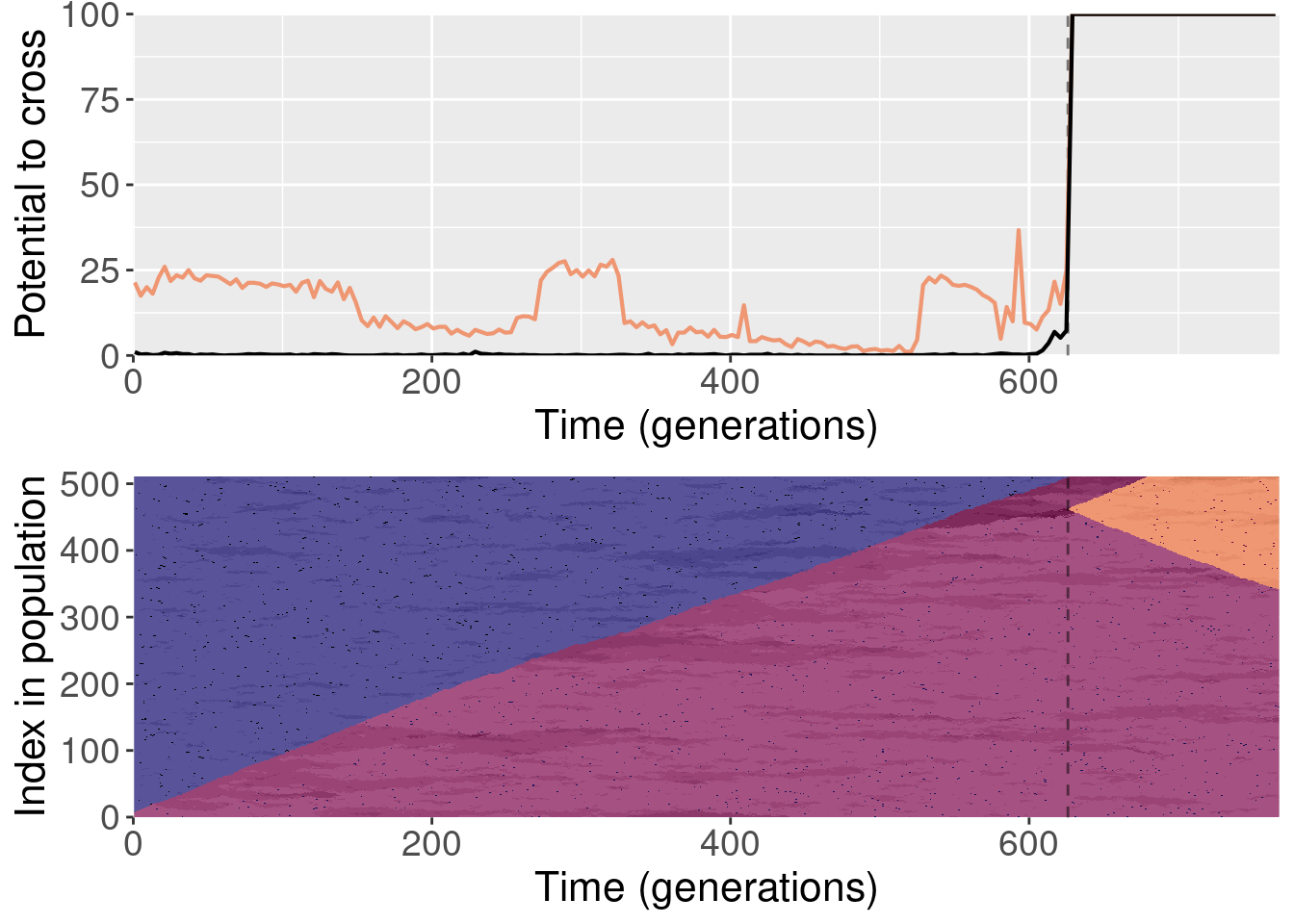
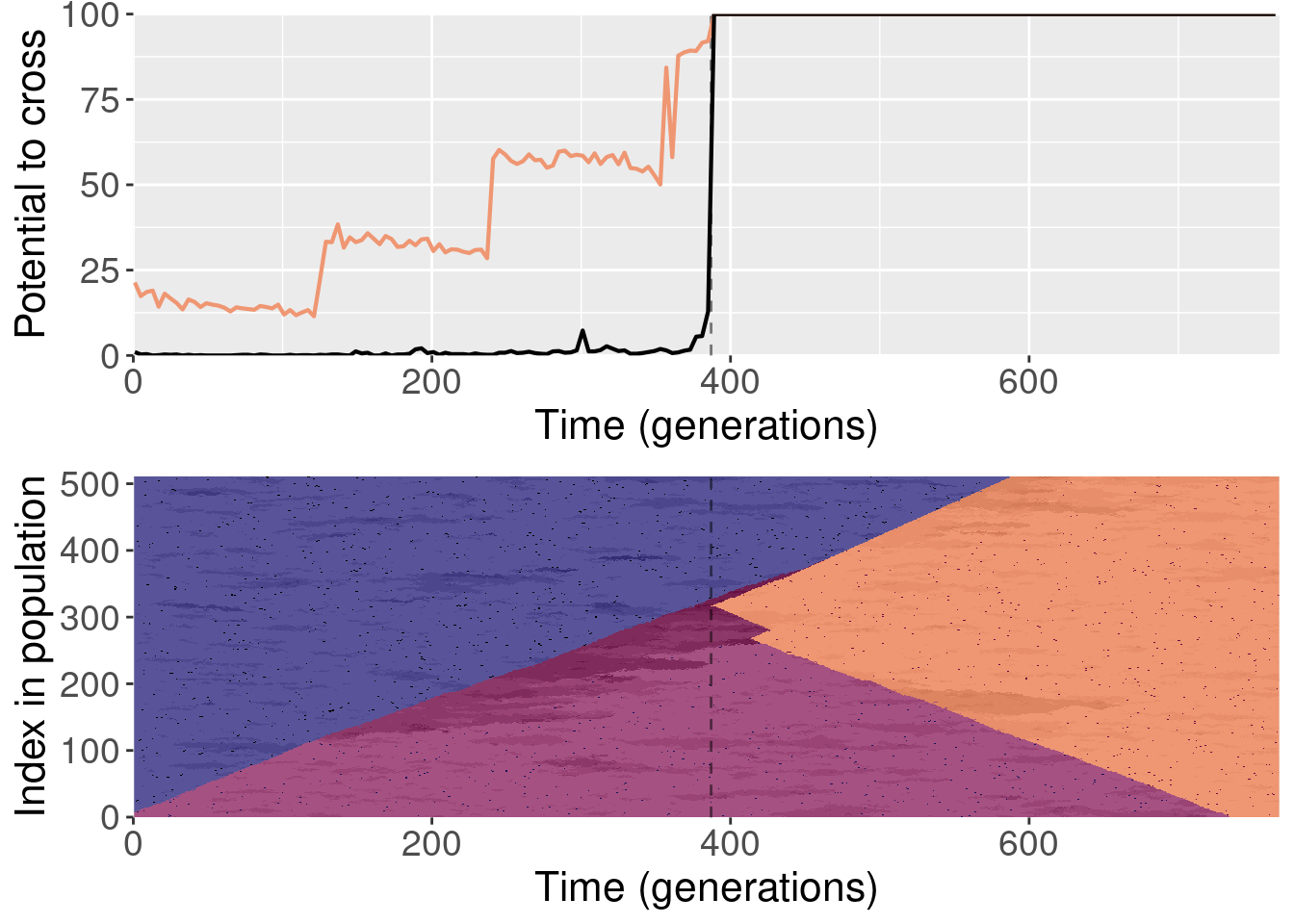
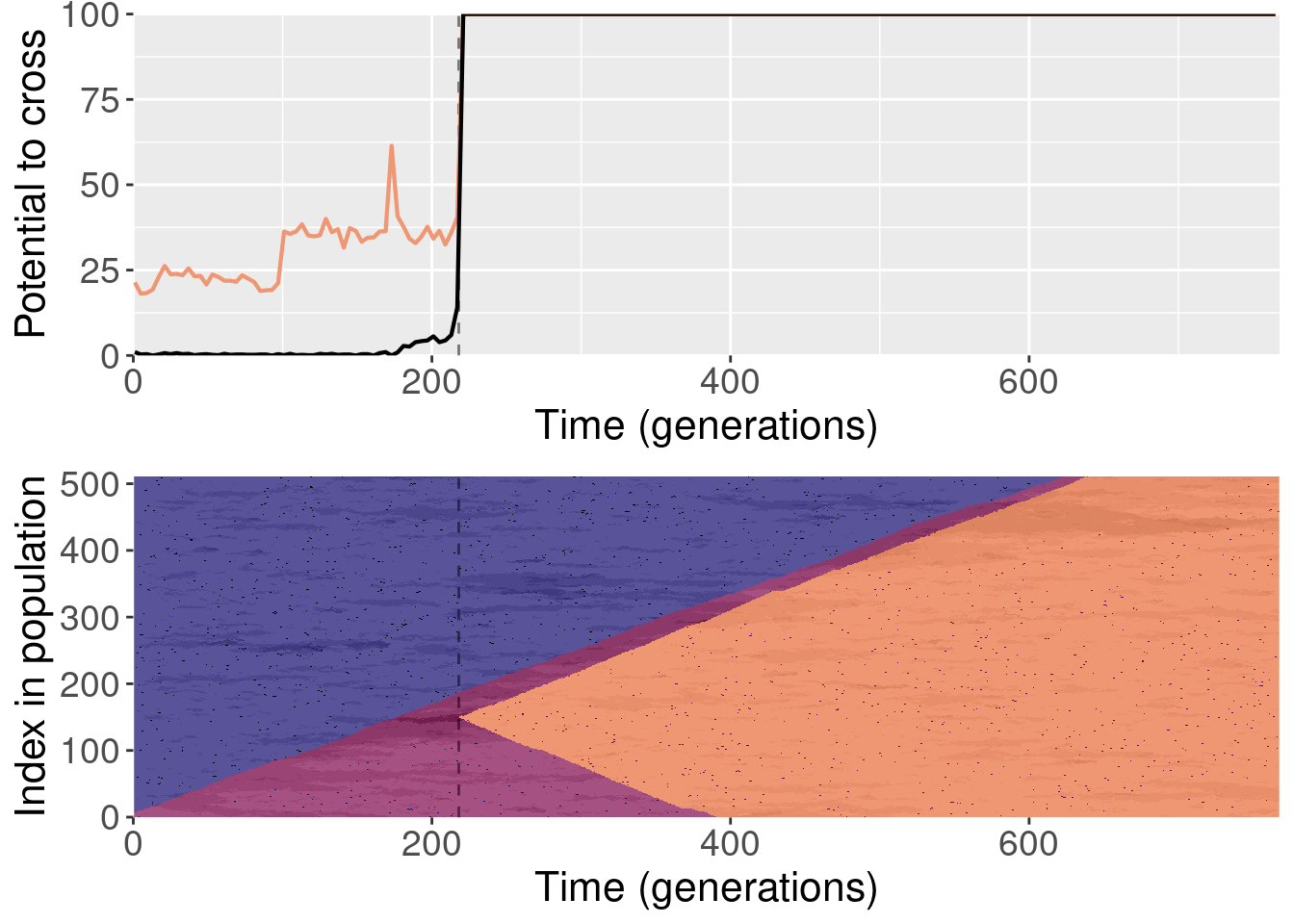
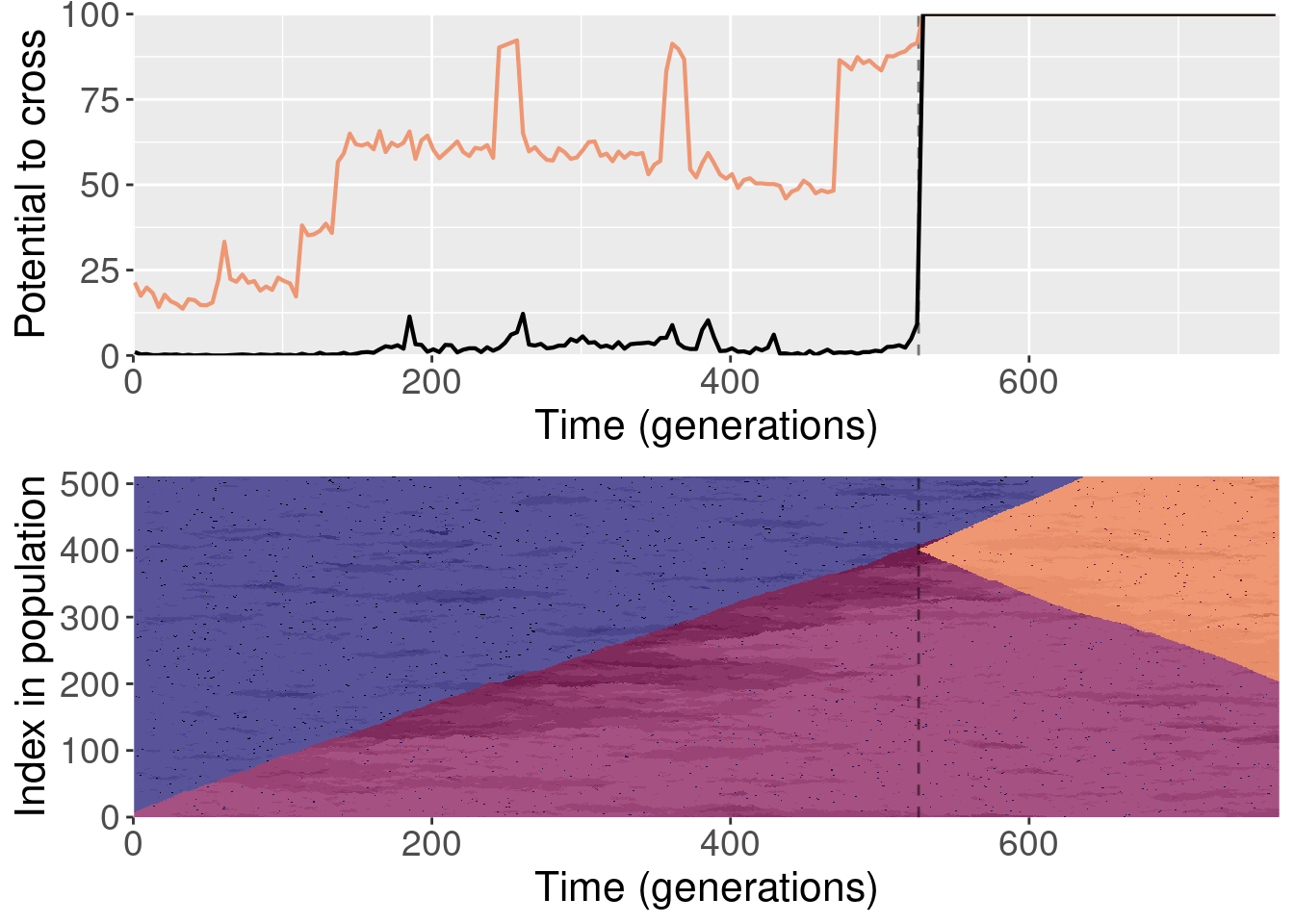
The paper shows one replay replicate that was shuffled prior to evolution. We actually ran 10 shuffled replays, and all of those files are included here.
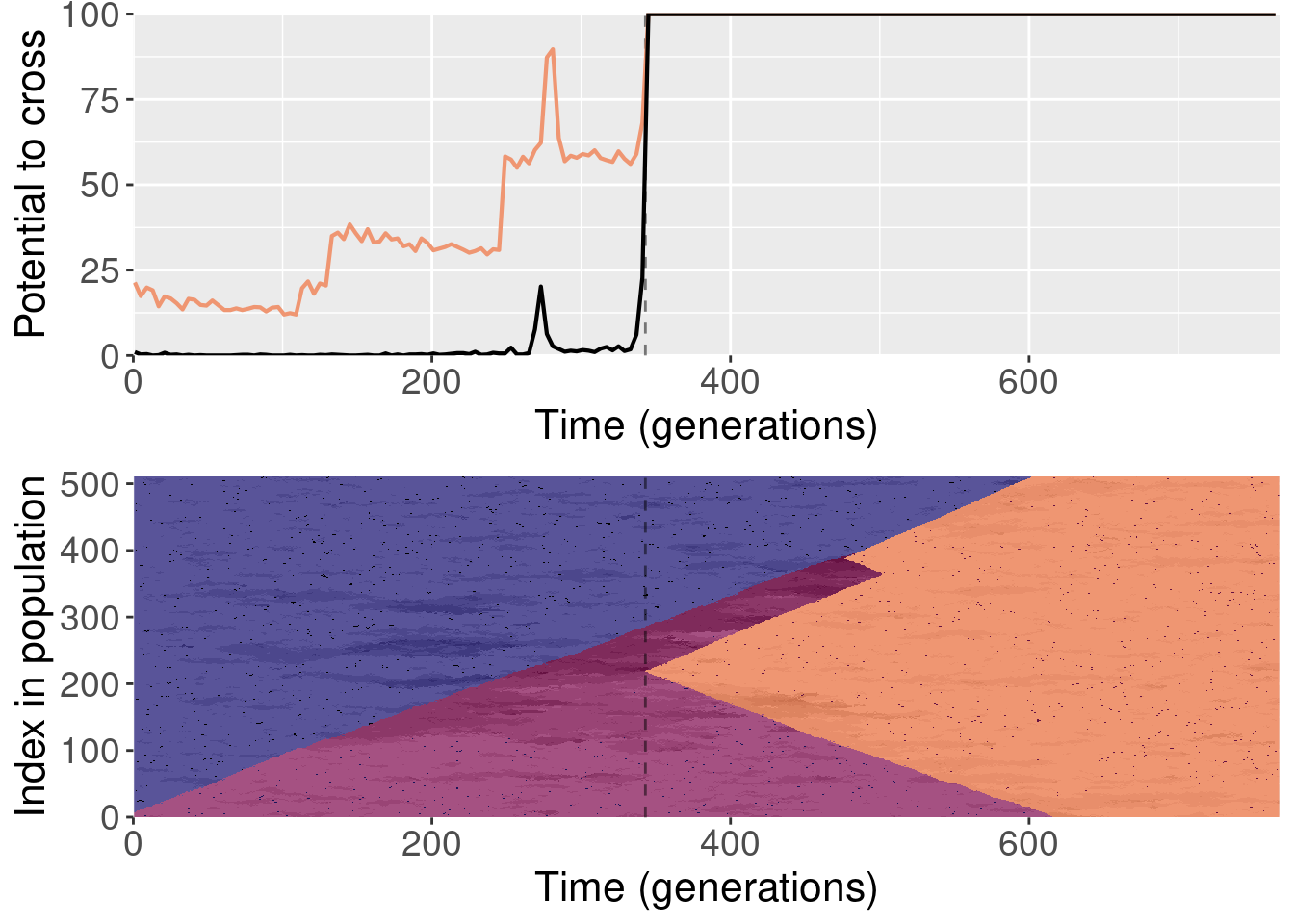
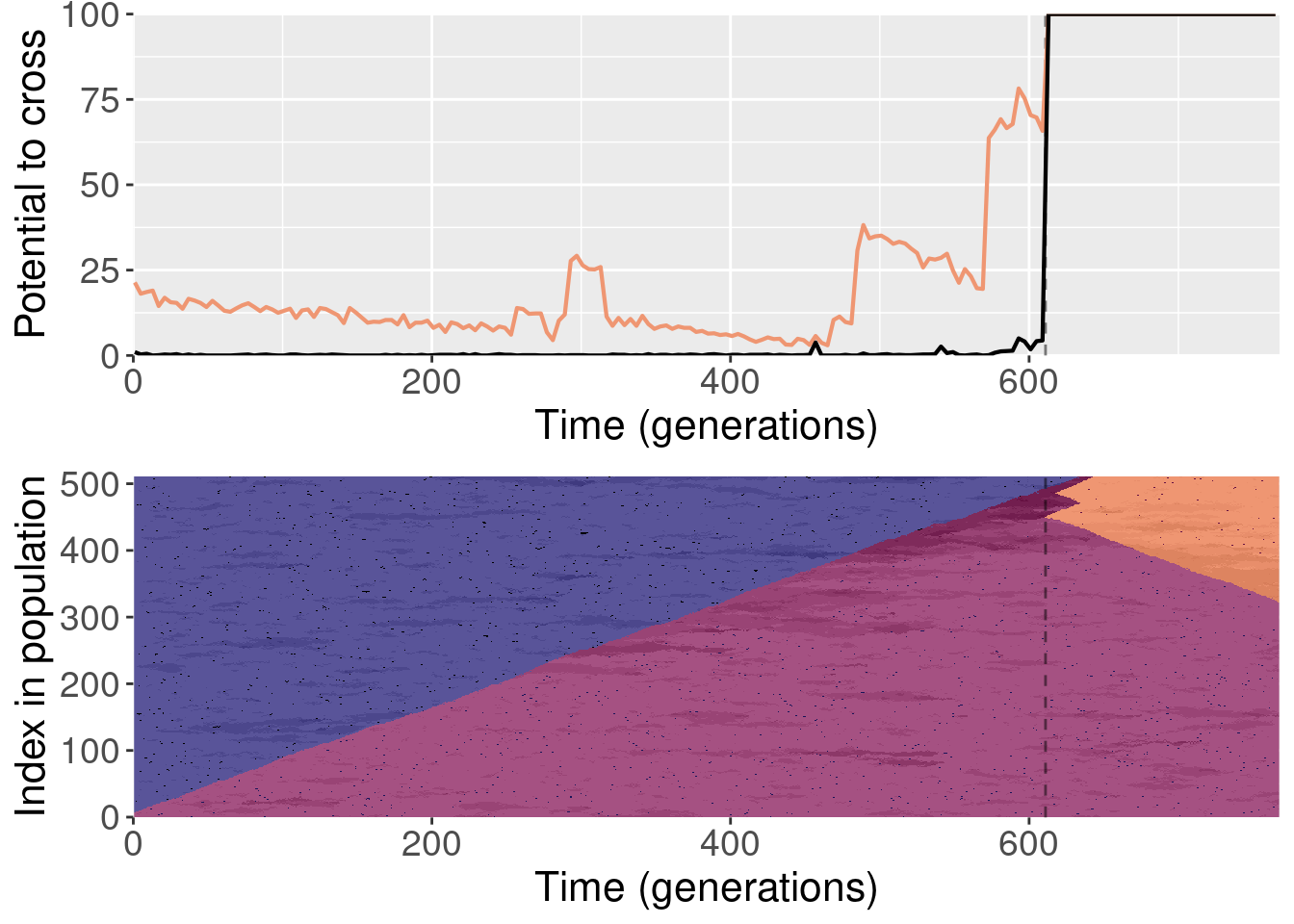
To clarify, for each replayed generation we took the population snapshot at that generation like normal. But for each replicate, we shuffled the organisms in the population prior to starting evolution. This destroyed any existing population structure, though new population structure could evolve again after.
The functions to generate the plots are not shown here.
They can be viewed in /experiments/2024_03_18_01__replays/analysis
For each plot, the top subplot shows the potential to cross with the normal replays (orange line) and the shuffled replays (black line). The bottom subplot shows the state of the population at each generation of the original replicate.
4.1 Dependencies
# External
library(ggplot2)
library(dplyr)
library(cowplot)
base_repo_dir = '../..'
exp_dir = paste0(base_repo_dir, '/experiments/2024_03_18_01__replays/')
# Internal
source(paste0(base_repo_dir, '/global_shared_files/global_analysis_variables.R'))